News
Inspiring the Next Generation of Scientists: Geneskool Field Trip at the GSC

The GSC was delighted to welcome a group of enthusiastic high school students as part of Geneskool, a Genome BC education program designed to spark interest in genomics and life sciences among students in grades 9 through 12.

Professor Steven Jones Elected Fellow of the Learned Society of Wales
The Genome Sciences Centre is proud to announce that our Co-Director and Head of Bioinformatics, Professor Steven Jones, PhD, FRSC, FCAHS, FRCP(Edin), FLSW, has been elected as a Fellow of the Learned Society of Wales—a prestigious recognition of excellence and leadership in academic research.

Enhancing Long-Read Sequencing: Introducing the Agilent Femto Pulse System at the GSC
We are excited to announce the arrival of the Agilent Femto Pulse System at the GSC! This high-sensitivity automated pulsed-field capillary electrophoresis instrument will be a game changer for nucleic acid quality control - detecting nucleic acids down to 50 fg/µL of DNA or RNA per sample and resolving fragments from 100 base pairs (bp) to over 165 kilobases (kb) in about 1.5 hours.
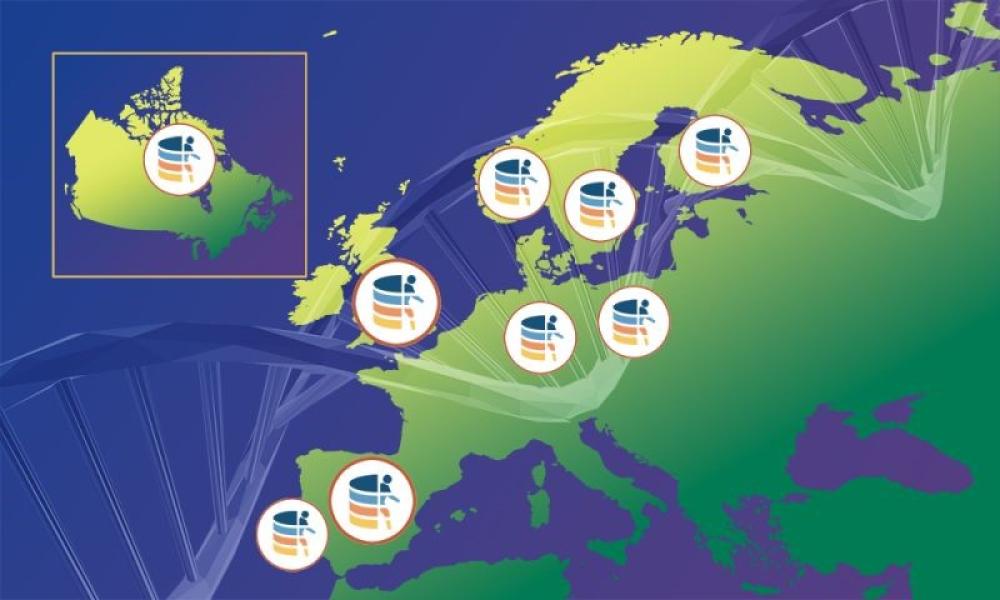
Canada joins Federated EGA, establishing a significant global repository for secure sharing of human genomic and health data
Canada’s Michael Smith Genome Sciences Centre at BC Cancer hosts the Canadian Node of the Federated EGA
In a major milestone for international biomedical research and the future of personalized medicine, the Canadian Genome-Phenome Archive (CGA), part of the Pan-Canadian Genome Library (PCGL) has joined the Federated European Genome-Phenome Archive (Federated EGA), marking the federation’s first significant expansion outside of Europe.
The CGA is hosted by Canada’s Michael Smith Genome Sciences Centre at BC Cancer.

POG and Dr. Marco Marra’s Killam Prize Featured in The Globe and Mail
Dr. Marco Marra and the groundbreaking Personalized OncoGenomics (POG) program have been prominently featured in The Globe and Mail following Dr. Marra’s recognition as a recipient of the 2025 Killam Research Prize in health sciences. This prestigious award highlights his contributions to Canadian research, particularly in the realm of genomics and precision cancer medicine.

GSC to Collaborate on Five Projects for the Canadian Precision Health Initiative
The GSC is proud to announce its collaboration on five groundbreaking projects as part of the Canadian Precision Health Initiative (CPHI). This ambitious national effort, launched today in Ottawa by the Honourable François-Philippe Champagne, Minister of Innovation, Science and Industry, represents an $81-million investment from the Government of Canada through Genome Canada.

Dr. Marco Marra appointed as UBC Killam Professor
UBC has announced the appointment of Dr. Marco Marra as one of three new University Killam Professors. The University Killam Professorship—the highest honour UBC can confer on a faculty member—recognizes exceptional teachers and researchers who are leaders in their fields, and who have received international recognition for their achievements.
Marco Marra
Department of Medical Genetics, Faculty of Medicine & Michael Smith Laboratories

Dr. Marco Marra appointed Officer of the Order of Canada
Her Excellency the Right Honourable Mary Simon, Governor General of Canada, announced today the appointment of Dr. Marco Marra as an Officer of the Order of Canada.

Laura Evgin selected Canada Gairdner Early Career Investigator
Congratulations to Dr. Laura Evgin for being selected as one of the 2025 Canada Gairdner Early Career Investigators for her work in CAR T-cell therapy.