Precision oncology—the use of tumour genomics to identify cancer cell vulnerabilities—is a rapidly evolving field of study. A new publication from BC Cancer summarizes how whole genome and transcriptome analysis can impact treatment options and clinical management for patients enrolled in BC Cancer’s Personalized OncoGenomics Program.
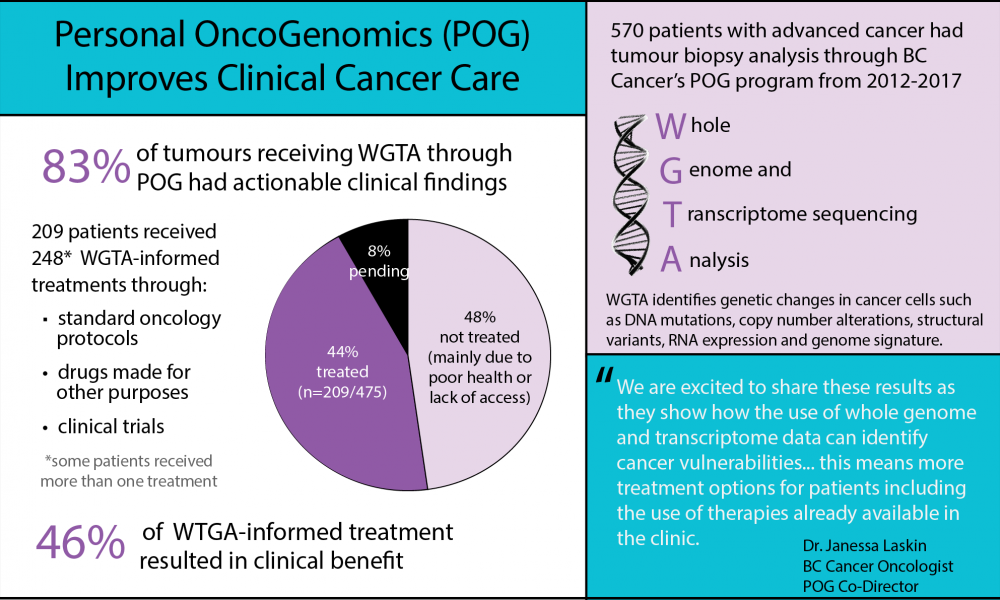
The Personalized OncoGenomics Program (POG), co-led by Drs. Janessa Laskin and Marco Marra, is a patient-centric research initiative that uses genomic data to study cancer growth, metastasis and response to treatment. A publication in Annals of Oncology presents clinical data from 570 adult patients with advanced or metastatic cancer enrolled in the program from 2012-2017, the largest report on POG clinical findings to date.
Together with previous published results on the genomic landscape of these tumours in Nature Cancer, researchers at Canada’s Michael Smith Genome Sciences Centre have shown the feasibility of considering comprehensive molecular data in the care of advanced cancer patients and the clinical implications illuminated through its use in a clinical cancer care setting.
Whole genome and transcriptome analysis (WGTA) of a biopsy sample from each patient’s tumour was conducted at Canada’s Michael Smith Genome Sciences Centre. Normal genomic DNA was also sequenced. WGTA used combined DNA- and RNA-based data to generate information on DNA mutations, copy number alterations, structural variants, RNA expression and genome signatures.
WGTA identified clinically actionable findings in most cases
For each patient, results of POG-generated WGTA were presented at a Molecular Tumour Board meeting, which included oncologists, pathologists, bioinformaticians and genome analysts. Together with clinical and pathology information, the team identified potential targets for treatment based on genomic findings.
Of the 570 samples with complete WTGA, 83% had clinically actionable findings identified at the Tumour Board Meeting. Of these 475 patients, 209 received POG-informed therapy. Some of these therapies were accessed through clinical trials or off label use of approved drugs, but many were standard treatments that the clinician prioritized above other standard treatments based on the genomic findings.
Reasons for not receiving treatment—of those with a clinically actionable finding—included poor patient health due to disease progression and lack of access to the recommended therapy due to geographic location and/or cost.
DNA sequencing and RNA expression information equally valuable
Of the patients who received POG-informed therapy, nearly half experienced clinical benefit according to reports provided by their physician. The majority of the POG-informed treatments were those supported by more than one WTGA data type.
RNA expression data were the most commonly used, contributing to 67% of cases with clinically actionable findings. One quarter (25%) of the treatments were informed by RNA-based data only. In contrast, 14% of clinically actionable cases were determined by DNA mutation alone. There was no significant difference between RNA and DNA-based data in terms of which POG-based treatments were most likely to provide clinical benefit.
"This was a striking result for us," notes scientist Dr. Erin Pleasance, one of the lead authors on the study. "From the beginning of POG, we integrated RNA expression information into our interpretation of the tumour biology, even though the vast majority of clinical molecular tests rely on only DNA or protein. The results of this study confirmed for us the added value of these data for informing treatment choices."
Patients access beneficial treatments
Patients accessed treatment through standard oncology protocols (46%), clinical trials (19%) and off-label use (35%). There was no difference in clinical benefit based on these three categories of treatment.
“The use of WGTA to inform funded standard therapy choices was an important approach, given issues around drug access and cost,” says Dr. Pleasance.
Study Implications
“We are excited to share these results as they show how the use of whole genome and transcriptome data can identify cancer vulnerabilities not detectable with standard DNA testing; this means more treatment options for patients including the use of therapies already available in the clinic,” says BC Cancer oncologist Dr. Janessa Laskin.
The feasibility and success of POG's WGTA approach has also supported expansion of pediatric cancer sequencing across Canada through PROFYLE and of a variety of projects as part of the Marathon of Hope Cancer Centres Network, of which BC Cancer is a core member. Combined, these efforts can expand our understanding of tumour biology, support the development of therapies and clinical trials, and ultimately improve health outcomes for patients living with cancer.
Learn More:
Learn more about BC Cancer Research, the POG program and Canada’s Michael Smith Genome Sciences Centre, where WGTA analysis for this study was conducted.
Acknowledgements:
The authors acknowledge the participation of patients and families, the POG team and Canada’s Michael Smith Genome Sciences Centre technical platforms. Data for this study was obtained, in part, from The Cancer Genome Atlas. Funding was provided by the BC Cancer Foundation, and their donors, and Genome British Columbia. Infrastructure costs were supported by Genome Canada, Genome BC, Canada Foundation for Innovation, the BC Knowledge Development Fund and the Canada Research Chairs program.
Citation:
Pleasance E, Bohm A, Williamson LM, Nelson JMT, Shen Y, Bonakdar M, Titmuss E, Csizmok V, Wee K, Hosseinzadeh S, Grisdale CJ, Reisle C, Taylor GA, Lewis E, Jones MR, Bleile D, Sadeghi S, Zhang W, Davies A, Pellegrini B, Wong T, Bowlby R, Chan SK, Mungall KL, Chuah E, Mungall AJ, Moore RA, Zhao Y, Deol B, Fisic A, Fok A, Regier DA, Weymann D, Schaeffer DF, Young S, Yip S, Schrader K, Levasseur N, Taylor SK, Feng X, Tinker A, Savage KJ, Chia S, Gelmon K, Sun S, Lim H, Renouf DJ, Jones SJM, Marra MA, Laskin J. Whole genome and transcriptome analysis enhances precision cancer treatment options. Ann Oncol. 2022 Jun 9:S0923-7534(22)01723-9. doi: 10.1016/j.annonc.2022.05.522. Epub ahead of print. PMID: 35691590.