More information is emerging on the role Fusobacterium nucleatum plays in tumor initiation, progression and treatment resistance. A recent study published in mBio reports on the pathogenesis of three different strains of the bacteria on two different host cells at the transcriptional and epigenetic level.
Fusobacterium nucleatum is a type of bacteria commonly found in the mouth that can spread to other parts of the body. Colonization of the pathogen is associated with several inflammatory conditions such as gum disease and inflammatory bowel disease.
F. nucleatum is also one of a growing number of infectious agents known to be involved in the cancer pathway. Research done in the laboratory of Dr. Robert Holt helped to specifically link F. nucleatum to colon cancer over ten years ago by showing the bacteria was more common in colon cancer cells than control cells through RNA sequencing. More recently, F. nucleatum has been linked to chemotherapy resistance and the metastatic spread of breast cancer.
Analysis of transcriptional and epigenetic host cell responses.
To explore host cell changes that occur in response to F. nucleatum, a team lead by Dr. Robert Holt performed transcriptome sequencing (RNA-seq) and chromatin immunoprecipitation sequencing (ChIP-seq) on two cell types exposed to three different bacterial strains. The results outlined in the paper’s abstract are captured in the figure below.
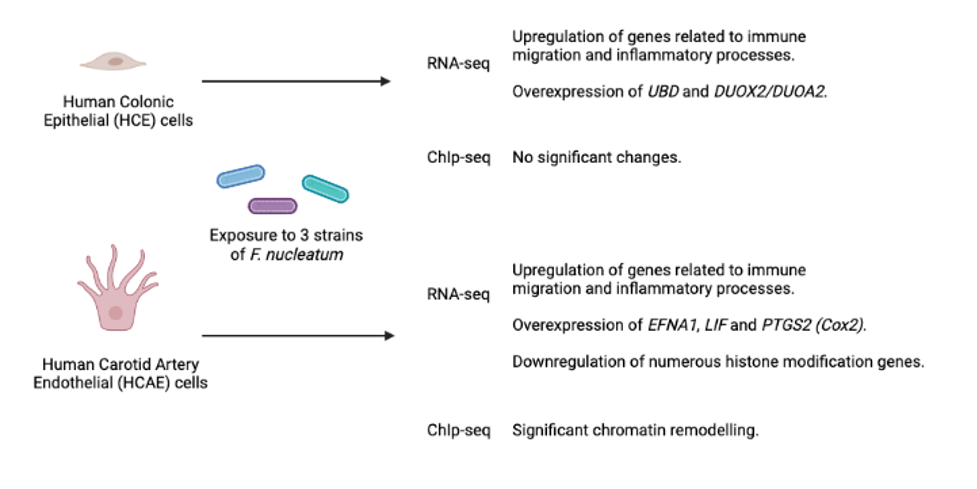
F. nucleatum upregulates genes related to immune migration and inflammatory processes.
Of note is the fact that increased inflammation and chemokine gene expression was seen in both host cell types. This is consistent with previous reports and suggests that this is a key aspect of host response to F. nucleatum that is highly conserved.
"There are a number of intriguing genes that were identified for both the conserved response to F. nucleatum and the cell line/tissue context specific response,” says Cody Despins, co-author of the study. “Hopefully further evaluation of the candidate genes identified here will give new insights into the mechanisms of F. nucleatum pathogenesis and its link to cancer."
Epigenetic changes are host cell specific.
For the paper’s other co-first author, Dr. Scott Brown, the most interesting outcome of this research was the detection of broad epigenetic changes in one of the F. nucleatum-exposed cell lines. “We only thought to directly measure these epigenome-wide histone changes after seeing downregulation of histone modification genes as a consistent change in host cells in response to F. nucleatum exposure.”
The fact that epigenomic changes vary between HCE and HCAE cells highlights the importance of considering both strain and cell type when exploring the role of F. nucleatum in cancer development.
“This represents a new lead in the search to discover how F. nucleatum may be dysregulating host cells and leading to cancer,” says Dr. Brown.
Acknowledgments
This reserach was supported by the Canadian Cancer Society, the Canadian Institutes of Health Research (CIHR), and Cancer Research UK (CRUK).
Image created with BioRender.com
Learn More
For a brief overview on oncomicrobes see Metagenomics - Infectious agents in Cancer.
For a more in-depth review of Fusobacterium and Colon Cancer, go to A Surprising Link Between Bacteria and Colon Cancer from Time Magazine.
Learn more about work being done by the Holt Lab.
Read about Grand Challenge funding from Cancer Research UK.
Citation
Despins CA, Brown SD, Robinson AV, Mungall AJ, Allen-Vercoe E, Holt RA. Modulation of the Host Cell Transcriptome and Epigenome by Fusobacterium nucleatum. mBio. 2021 Oct 26;12(5):e0206221. doi: 10.1128/mBio.02062-21. Epub 2021 Oct 26. PMID: 34700376; PMCID: PMC8546542.
*bold font indicates members of the GSC.