Lung cancer is consistently within the top three cancers in terms of prevalence worldwide. Many therapies directly target and inhibit key proteins in cellular pathways, though patients often develop resistance to such treatments. To identify other protein targets for therapies, the labs of the GSC’s Dr. Gregg Morin and Integrative Oncology’s Dr. William Lockwood worked to characterize a small molecule compound, LCS3, and demonstrate its potential use by treating lung cancer cells.
Lung cancer and therapies
Lung cancer—more specifically, human lung adenocarcinoma—can occur as a result of specific mutations within critical proteins (e.g., epidermal growth factor receptor, EGFR) that function in key cellular pathways. A therapy directly targeting and inhibiting the activity of mutant proteins in cellular pathways critical for cancer cell development and survival is one successful avenue for treating lung adenocarcinoma.
Unfortunately, treated patients often end up developing resistance to targeted therapies. Fortunately, other proteins—not necessarily those directly involved in the aberrant cellular pathway—can be selected as alternative targets (e.g., proteins that assist in the growth and survival of the lung cancer cells).
Phenotype-based screening of small molecule compounds
One approach to targeting and inhibiting a mutant protein is to use a small molecule compound (i.e., a drug) that binds to the target protein/enzyme. When a susceptible target protein is known, candidate small molecule inhibitors are typically identified using in vitro screens to assay a library of compounds for their ability to inhibit the activity of the protein, usually an enzyme. Alternatively, phenotype-based screening, also termed cell-based screens, of a compound library can be used to identify compounds that produce a desired cellular phenotype response. LCS3, a small molecule compound, was identified using this strategy.
Compounds can inhibit enzyme function in various ways. If the compound binds to the target enzyme’s active site (a specific region present in all enzymes)—like a key within a lock—this prevents the enzyme from binding its regular target, the substrate, and prevents the enzyme from carrying out its normal function. This specific mechanism is called competitive inhibition as the compound is directly competing with the substrate for binding at the enzyme’s active site. If the compound instead binds to the enzyme-substrate complex—where the enzyme is already bound to the substrate—and then prevents the complex from functioning, this type of inhibition is called uncompetitive inhibition.
To develop screening hits into drugs, further chemical modifications of the initial hit are usually needed to improve the efficacy, selectivity, and in vivo biological performance. This is greatly enabled when the target protein and its functional pathway are known. Thus, to develop a phenotype screen hit into a drug candidate, it is necessary to fully characterize a compound’s specific protein target and mechanism of action through additional experiments. However, identifying the specific protein targeted by a compound can be challenging.
Characterizing the LCS3 compound
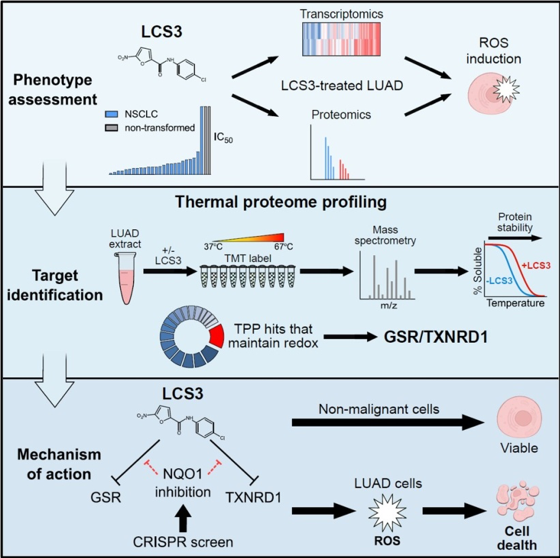
Fraser Johnson, lead author of the study and co-supervised by the GSC’s Head of Proteomics, Dr. Gregg Morin, and Integrative Oncology’s Dr. William Lockwood, worked alongside colleagues from the GSC, the U.S.A., and Germany, in research published in Cell Reports. In the study, the researchers characterized a previously identified small molecule compound, LCS3, that was found to potently impair the growth of human lung adenocarcinoma cells in a phenotype screen published in 2009, but whose target and mechanism of action proved difficult to determine.

The researchers found that this compound induced oxidative stress and lethality in lung adenocarcinoma cells. To identify the protein target of LCS3, the researchers used thermal proteome profiling (TPP)—a mass spectrometry-based proteomics technique that exploits a protein’s altered stability at high temperatures based on whether or not it is bound to a compound.
From their experiments, they identified two disulfide reductases—GSR and TXNRD1—as the specific targets of LCS3 and confirmed that LCS3 inhibits disulfide reductase activity through an uncompetitive mechanism, and leads to oxidative stress. Their research characterizing LCS3 shows the potential therapeutic benefit of inhibiting disulfide reductases in human lung adenocarcinoma cells and, more generally, demonstrates that TPP can be used to identify specific targets of small molecule compounds.
Acknowledgements:
This study was supported financially by the BC Cancer Foundation, the Cancer Research Society, the Terry Fox Research Institute, and the Canadian Institutes of Health Research.
Learn more:
Learn more about the ongoing research by the Gregg Morin Lab at the GSC.
Learn more about the discovery of the LCS3 compound here where it was identified by Somwar et al., (2009).
Citation:
Johnson FD, Ferrarone J, Liu A, Brandstädter C, Munuganti R, Farnsworth DA, Lu D, Luu J, Sihota T, Jansen S, Nagelberg A, Shi R, Forcina GC, Zhang X, Cheng GSW, Spencer Miko SE, de Rappard-Yuswack G, Sorensen PH, Dixon SJ, Guha U, Becker K, Djaballah H, Somwar R, Varmus H, Morin GB, Lockwood WW. Characterization of a small molecule inhibitor of disulfide reductases that induces oxidative stress and lethality in lung cancer cells. Cell Rep. 2022 Feb 8;38(6):110343. doi: 10.1016/j.celrep.2022.110343. PMID: 35139387.
*bold font indicates members of the GSC.