
In a collaboration between the GSC’s Bioinformatics Technology Lab, the Michael Smith Lab at UBC, NC State University in Raleigh and Laurentian Forestry Centre Natural Resources Canada, the nuclear and mitochondrial genomes of Pissodes strobi (an insect also known as the spruce weevil or the white pine weevil) were sequenced. The results could help researchers control the destructive impact of this forest pest.
It might be a small insect, but Pissodes strobi (referred to here as the spruce weevil) has a huge impact. Part of the beetle family, the spruce weevil—one of more than 60,000 species in the taxon Curculionidae (or “true weevils”)—is highly destructive to North American forests.
Every spring, adult female spruce weevils lay eggs at the tip of tree leaders (shoots that have grown in the last two years). When the larvae hatch, they feed downward—damaging the leader.
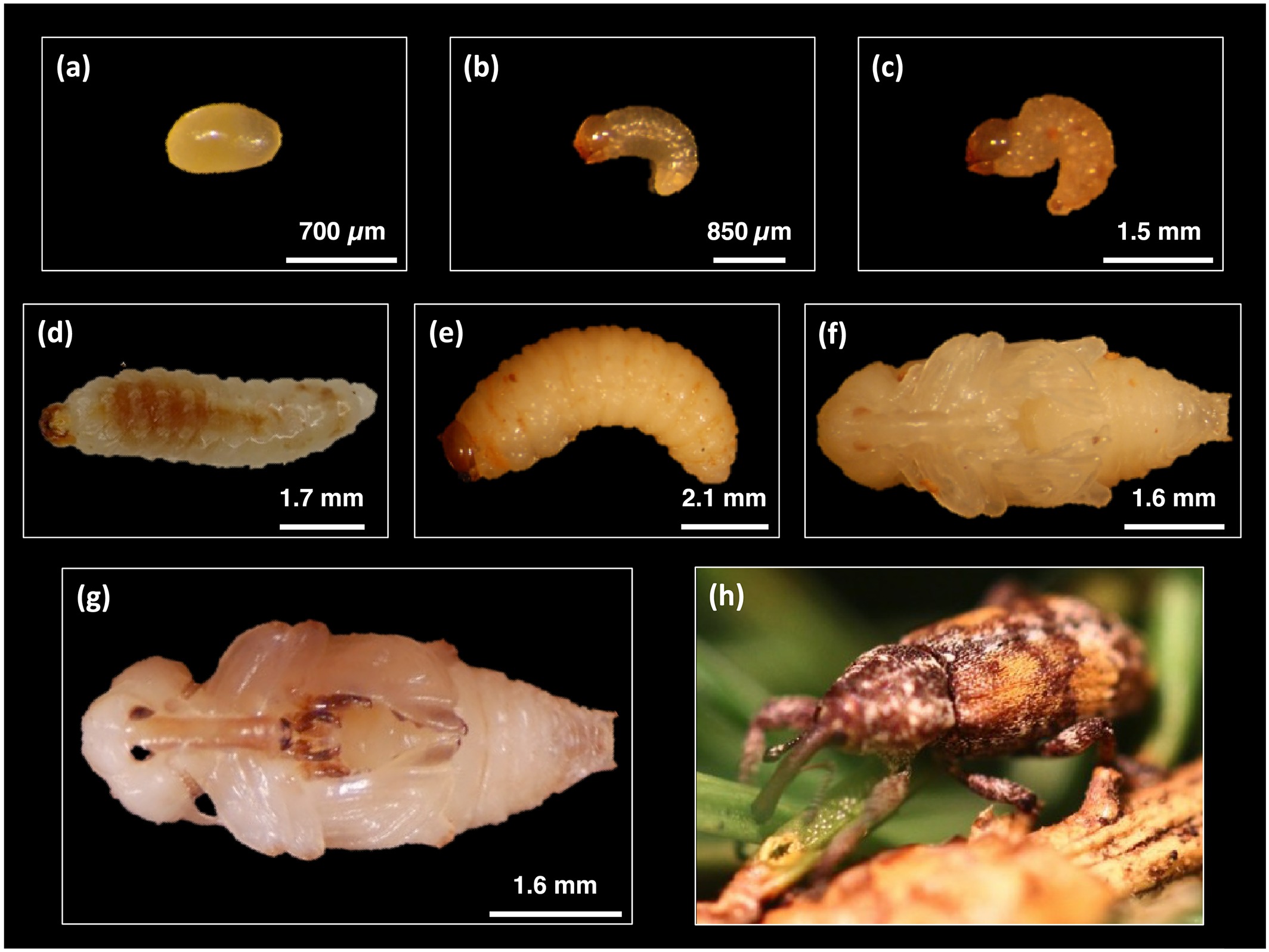
Trees damaged by spruce weevils become forked and damaged. Over time, repeated weevil attacks cause a loss of growth, reduction in wood quality and tree death. In BC, the spruce weevil is a threat to several commercially important conifer species including Sitka spruce, Engelmann spruce and white spruce.
Sequencing the Spruce Weevil Genome
The nuclear and mitochondrial genome of spruce weevil was sequenced as part of CanSeq150—an initiative to commemorate the 150th Birthday of Canada in 2017. The aim of this collaborative program, launched by CGEn, Canada’s national platform for genome sequencing and analysis, is to sequence 150 new genomes to support sequence-based genomics research in Canada and inform future research in areas such as biodiversity and conservation.
“Assembling and analyzing the genome of the spruce weevil has been very exciting for me,” says Kristina Gagalova, PhD Candidate in the Birol lab and co-first author of the study. “Its genome size is one of the largest sequenced so far when compared to related species.”
Results were published in G3: Genes, Genomes, Genetics with two notable findings.
First, the team discovered sequences interspersed among the spruce weevil DNA sample that most closely resemble Wolbachia, a type of bacteria. This suggests that Wolbachia is an endosymbiont—bacteria that forms symbiotic relationships with other species—of the spruce weevil. The bacterial sequencing reads were assembled separately and released as a microbe genome.
The second major finding was substantial expansion within the spruce weevil nuclear genome, relative to other members of the weevil taxon, possibly driven by an abundance of transposons. Although more analysis is required, this repeat expansion is likely driving the weevil’s diversity.
“By releasing the spruce weevil genome we provide a reference genome for one of the most disruptive conifer pests in North America,” says Kristina. “I hope this work can trigger further research about widespread spruce weevil populations and how they impact conifers in different environments.”
Acknowledgements:
Funding was primarily provided by individuals recognized at http://canadiana.bcgsc.ca, the Canadian Foundation for Innovation and CGEn CanSeq150 program.
Weevil Photo Credit: Justin G. A. Whitehill, North Carolina State University, USA
Learn more:
Learn more about how the weevil is affecting BC Forests and the difference between the transcriptomes of weevil-resistant and susceptible Sitka spruce.
Learn more about the GSC, the Bioinformatics and Technology Lab and CanSeq150.
Citation:
Kristina K Gagalova, Justin GA Whitehill, Luka Culibrk, Diana Lin, Véronique Lévesque-Tremblay, Christopher I Keeling, Lauren Coombe, Macaire MS Yuen, Inanç Birol, Jörg Bohlmann, Steven JM Jones. The genome of the forest insect pest Pissodes strobi reveals genome expansion and evidence of a Wolbachia endosymbiont. G3: Genes, Genomes, Genetics PMID: 35171977
*bold font indicates members of the GSC.