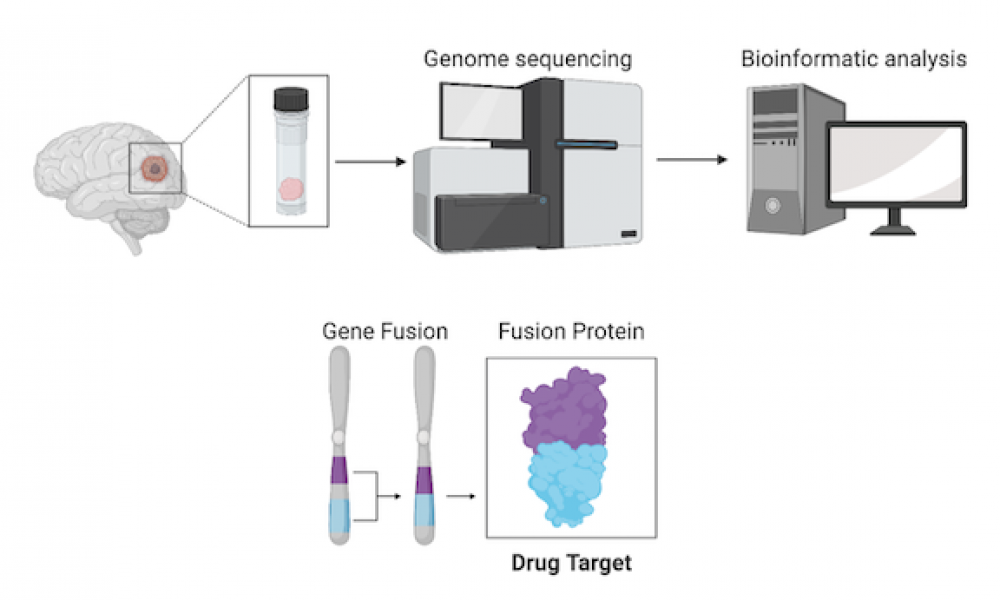
Whole genome sequencing allows for the identification of novel genomic alterations that are not detected by standard clinical techniques. In some cases, identification of these features can reveal therapeutic options for patients with otherwise untreatable cancers.
In a new study published in the Wiley journal Clinical Case Reports, researchers presented a case in which whole genome analysis unveiled a drug target for a pediatric patient with glioblastoma, a rare and often fatal form of brain cancer.
The study, led by Dr. Christopher Dunham, Clinical Associate Professor in the Department of Pathology and Laboratory Medicine at the University of British Columbia, was part of the Personalized OncoGenomics (POG) program at BC Cancer—a precision oncology initiative that performs whole genome and transcriptome sequencing to identify targetable genetic alterations in patients with advanced disease.
Pediatric brain tumours are the leading cause of childhood cancer death. The prognosis for children with glioblastoma, which accounts for as few as 2.5 per cent of primary pediatric central nervous system tumors, remains very poor, with a median overall survival of nine to 15 months.
“Novel therapeutic options for children with brain tumours are desperately needed,” says Dr. C. Dunham. “Whole genome and transcriptome sequencing allows us to detect genomic alterations that we otherwise would not have looked for. Some of these may be completely novel, providing researchers with an avenue for drug development, while others may be common in other cancers and may be treatable with currently available therapeutics.”
Whole genome analysis of a pediatric glioblastoma patient enrolled in the POG program led to the discovery of a gene fusion—a genomic alteration where two previously independent genes become fused together—involving the gene NTRK2. This type of fusion has been implicated in several cancer types and clinical trials of targeted therapies in both children and adults have yielded promising results.
In addition to the NTRK2 fusion, transcriptomic analysis uncovered another potential treatment avenue. Expression of the gene PD-L1 was found to be upregulated in the tumour tissue, suggesting potential susceptibility to immune checkpoint inhibitors.
“These findings demonstrate the value of whole genome and transcriptome analysis for personalized cancer care by identifying potential existing or novel medical treatment strategies where, in some cases, therapeutic options are limited,” says Dr. Heidi Britton and first author on the study. “The findings also contribute valuable information to the body of knowledge on childhood brain tumors.”
The authors of the study gratefully acknowledge the participation of the patient and are also grateful for the funding support provided by the BC Cancer Foundation.
Learn more about the Personalized OncoGenomics program.
Learn more about Cancer Genomics.
Reference:
Heidi M. Britton, Adrian B. Levine, Yaoqing Shen, Karen Mungall, Jonathan Serrano, Matija Snuderl, Erin Pleasance, Steven J. M. Jones, Janessa Laskin, Marco A. Marra, Shahrad R. Rassekh, Rebecca Deyell, Stephen Yip, Sylvia Cheng, Chris Dunham. NTRK2 Fusion driven pediatric glioblastoma: Identification of oncogenic Drivers via integrative Genome and transcriptome profiling. Clinical Case Reports. 2021;00:1–6.