Acute myeloid leukemia (AML) is an aggressive cancer that manifests in the bone marrow and affects the production of blood cells. The rapid progression of the disease, combined with the large degree of heterogeneity between AML patients, complicates individualized treatment planning.
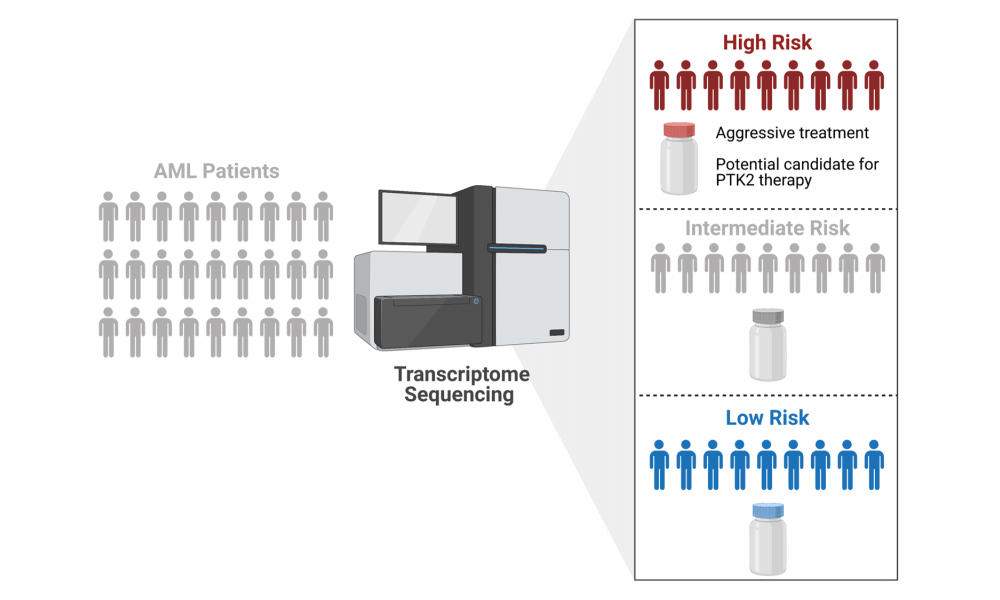
In a new study published in Nature Communications, led by GSC Distinguished Scientist Dr. Aly Karsan, researchers used the power of next generation sequencing (NGS) technologies to develop a clinical test that improves patient stratification. Clinical implementation of the test will allow clinicians to better determine which of their patients fall into high-, intermediate- and low-risk categories, enabling personalized treatment planning.
Current methods for the stratification of AML patients into different risk groups results in approximately half of patients being classified as intermediate-risk. However, there is extensive genetic heterogeneity within this group and assigning an appropriate treatment regimen for each patient remains a significant challenge.

“AML is a very rapidly acting cancer. Understanding which patients are likely to respond to therapy, which patients are not likely to respond, and which patients would benefit form more aggressive upfront therapy, can have dramatic consequences for treatment outcomes,” says Dr. T. Roderick Docking, first author on the study.
To address this issue, Dr. Karsan’s research team set out to bring AML diagnostics and stratification into the NGS era. They found that, compared to whole genome and exome sequencing, a standalone RNA-Seq assay provided the greatest diagnostic benefit, allowing for the identification of expressed gene fusions, single nucleotide variants, short insertion and deletion mutations, and alterations in gene expression.
“We then we asked, how can we actually improve on the status-quo? We can tell whether these recurrent gene fusions and mutations are there or not, but how do we use the wealth of information in the transcriptome to actually better stratify patients,” says Dr. Docking.
Expression data from 154 AML patients were used develop an AML prognostics score, which was validated across four additional cohorts and, importantly, was found to be consistent for both adult and pediatric AML. The risk score allowed for the re-stratification of approximately one quarter of AML patients into their correct risk groups.
The researchers used the transcriptome data to further probe the high-risk AML group for potential therapeutic targets. They found that a subset of high-risk patients had increased expression of the gene PTK2, which encodes focal adhesion kinase—a protein targeted by drugs currently in clinical trials for other cancers. They then tested the drugs against PTK2-overexpressing cells in the laboratory and found that focal adhesion kinase inhibitors may be effective for the treatment of high-risk AML.
The RNA-Seq assay was developed using a clinical validation framework with the sensitivity and specificity that will be required for clinical implementation. Moving forward, the group is working with the GSC to establish the laboratory protocols and bioinformatic pipelines for clinical use.
T. Roderick Docking, Jeremy D. K. Parker, Martin Jädersten, Gerben Duns, Linda Chang, Jihong Jiang, Jessica A. Pilsworth, Lucas A. Swanson, Simon K. Chan, Readman Chiu, Ka Ming Nip, Samantha Mar, Angela Mo, Xuan Wang, Sergio Martinez-Høyer, Ryan J. Stubbins, Karen L. Mungall, Andrew J. Mungall, Richard A. Moore, Steven J. M. Jones, İnanç Birol, Marco A. Marra, Donna Hogge, Aly Karsan. A clinical transcriptome approach to patient stratification and therapy selection in acute myeloid leukemia. Nature Communications.
This work was supported by Genome BC, the BC Cancer Foundation through the Leukemia and Myeloma Program, the BC Provincial Health Services Authority, the Terry Fox Research Institute, and the Leukemia and Lymphoma Society of Canada.
Learn more about research in the Karsan lab.
Learn more about research at the GSC.