Researchers at Canada’s Michael Smith Genome Sciences Centre have developed an elegant method for tracking tissue samples through complex laboratory processes using unique DNA identifiers.
As precision medicine for the treatment of cancer and other diseases becomes a reality, there is increased demand for DNA sequencing. With the high volumes of samples now being sent for sequencing analysis, Scientists at Canada’s Michael Smith Genome Sciences Centre (GSC) have created streamlined, tightly regulated processes that minimize errors and ensure production of high quality data.
In a study published in the Journal of Molecular Diagnostics, GSC researchers describe a new method for keeping track of the thousands of samples sent to the Centre using DNA itself.
Every sample received by the GSC is processed through a series of steps, which together compose a “pipeline”. With up to 15 different pipelines running at any one time, and each consisting of many intricate steps, keeping track of samples as they move in parallel through these complex processes is no small feat.
“If we didn’t track our samples effectively, we wouldn’t be able to find or do anything,” says Dr. Richard Moore, Sequencing Group Leader at the GSC, “It is essential that we track each sample through whichever pipeline they enter.”
Through the pipeline
From the time a sample is received by the GSC to the time it is loaded onto a sequencer, it will have passed through several instruments and the hands of multiple laboratory technicians. High confidence in sample identity at each step - especially when processing clinical samples - is of utmost importance; multiple levels of control are in place to ensure that the data produced at the end of a pipeline correspond to the correct input sample.
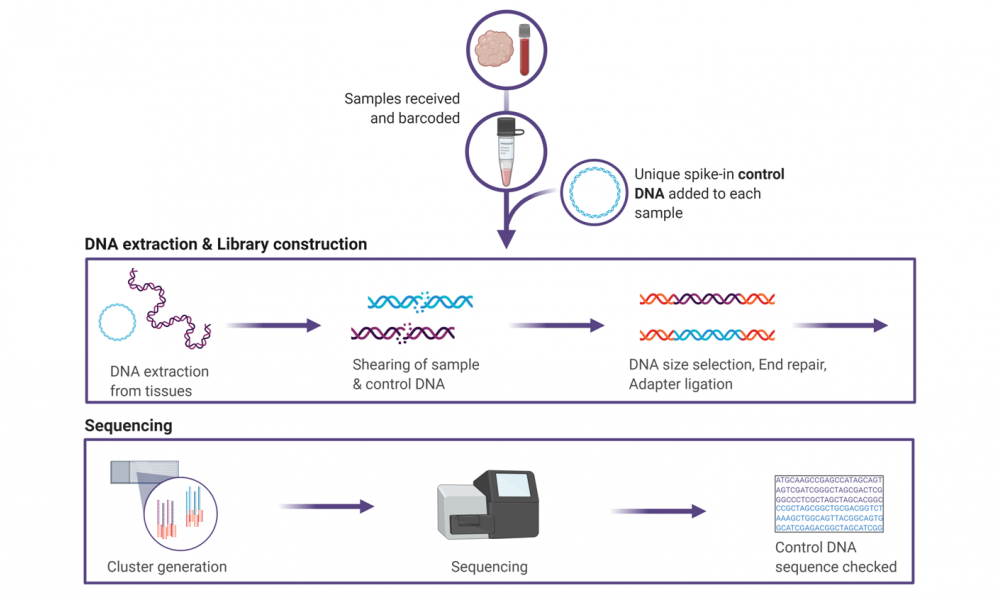
As soon as samples enter the doors of the GSC, they are provided with an optical barcode, allowing samples to be tracked using a Laboratory Information Management System, or LIMS. Every processing step and storage location is logged in the LIMS so that scientists know where each sample is at all times and what has been done to it.
Other tracking controls include having one technician observe another as samples are transferred between tubes and plates, checking specific regions of the DNA sequence called SNPs (pronounced “snips”) before and after samples have moved through the pipeline, and fusing unique DNA sequences onto the sample DNA before it is loaded onto the sequencers.
Ensuring quality
While these controls are very effective when used in combination, none can be used to definitively confirm sample identity at every stage of the pipeline, or be used to distinguish between multiple samples from the same patient.
“For some samples, even more controls are needed to ensure data quality,” says Dr. Moore, “For example, if you had multiple samples from the same person, or you are running twin studies, you can’t check which is which by doing SNP concordance assays.”
The novel method developed by Dr. Moore and scientists at the GSC relies on circular pieces of DNA called plasmids. Each of the approximately 5,000 different plasmids created at the GSC have a known, unique DNA sequence. As soon as a sample is received, before the tissues have even been broken open for DNA purification, a control plasmid is added.
As the sample proceeds through all of the processing steps of the pipeline, the control plasmid goes along with it. Once the sequencing data has been produced, technicians can ensure that the DNA sequence of the control matches the plasmid that was added at the beginning of the pipeline.
“The beauty of using plasmid DNA is that we can use bacteria to produce the plasmid again and again, so we have a never-ending supply of the same thing without needing to re-order more,” says Dr. Moore.
With so many levels of control, pipeline errors are extremely rare. But occasionally, the DNA sequence produced does not match what researchers expected. This method can be used to distinguish between a sample swap that may have happened somewhere along the pipeline, cross-contamination between samples run in parallel or an incorrect sample sent to the GSC from collaborators.
As we move closer to making precision medicine through genome sequencing the norm, the demand for parallel processing of large numbers of samples within high-throughput laboratories will continue to increase. Controls such as the one developed at the GSC will be essential to ensure the highest level of data quality and accuracy, providing patients and clinicians with high confidence diagnostics for personalized treatment planning.
The study was led by Dr. Aly Karsan. It was funded by Genome BC, BC Cancer Foundation and the Provincial Health Provincial Health Services Authority. The study was published in the Journal of Molecular Diagnostics.