
Although the human genome was first sequenced three decades ago, to this day, there are still DNA regions that have yet to be resolved (though we are getting very close to completion as of 2021). These missing DNA regions represent sequencing gaps that can arise due to inconsistent read coverage depth and/or repetitive DNA sequences, which are tricky to assemble without a reference sequence. Developed by researchers in Dr. Inanç Birol’s Lab, GapPredict uses deep learning to predict missing DNA bases in the unresolved regions of genome assemblies.
Gap-filling: an unresolved issue of de novo genome assembly
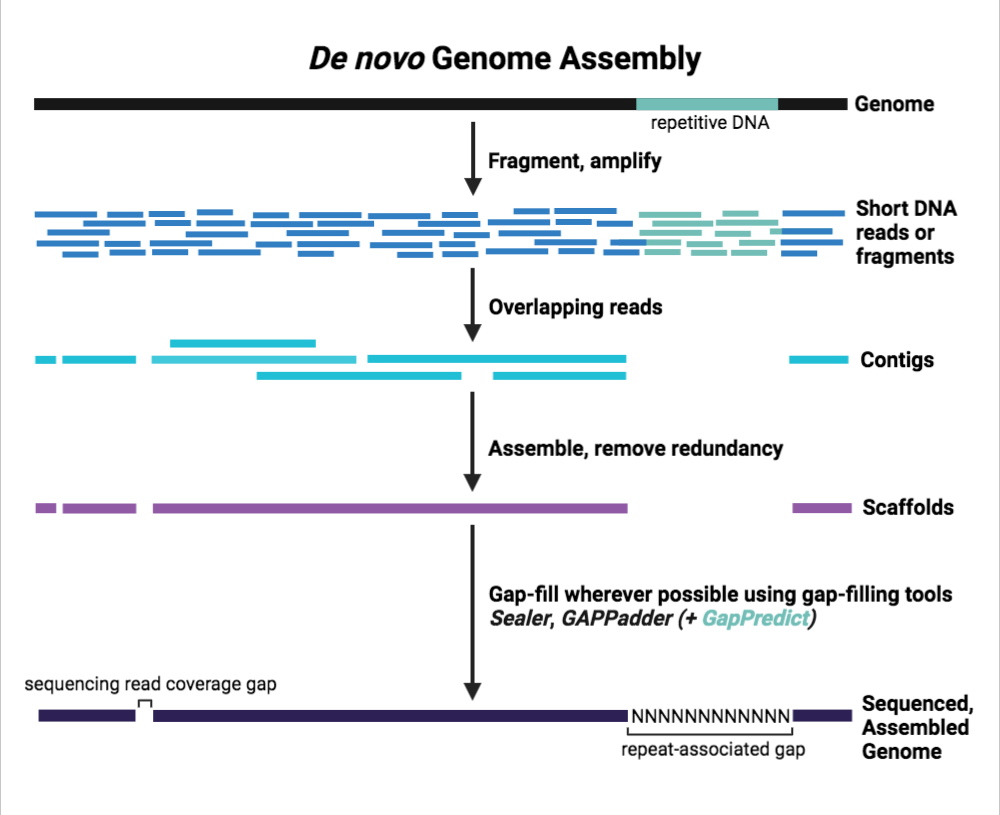
In order to sequence the human genome, several scientists used the whole genome shotgun approach. Several copies of the complete human genome were randomly fragmented, sequenced, and then reassembled using computer algorithms to align the overlapping DNA fragments. Genome assembly efforts ran into problems when faced with long, repetitive DNA sequences (e.g., it was unclear how long such repetitive regions were) and thus sequencing gaps, areas of the genome that cannot be resolved with certainty, were identified.
To this day, gap-filling remains a problem when it comes to de novo genome assembly, and this issue persists with the use of next generation sequencing (NGS) technology with short and long read sequencing.
GapPredict, a proof-of-concept model
To address the gap-filling problem, various algorithms and programs have already been developed and most use heuristic (quick, rigid, well-defined) approaches to predict and fill in DNA sequencing gaps. Unlike most previous gap-filling programs, GapPredict, developed by the Birol Lab and recently published in the journal, IEEE/ACM Transactions on Computational Biology and Bioinformatics, uses deep learning (slower, flexible, and evolving) to predict unresolved DNA in sequencing gaps.
GapPredict can complement gap-filling solutions of other tools
Deep learning, a subset of machine learning, is a process by which computers are fed features of a dataset to “teach” them how to make predictions about a new dataset. Reminiscent of human learning, deep learning models also require “training”. GapPredict exploits the large amount of information that represents the sequence context of gaps (e.g., the sequence preceding the gap) to conduct deep learning.
GapPredict represents a proof-of-concept model that applies deep learning to the problem of gap-filling and attempts to predict the most likely DNA sequence of such gaps. By benchmarking GapPredict against traditional gap-filling tools, researchers concluded that while GapPredict may not be suitable for large-scale application, it was in fact capable of filling in gaps left by traditional gap-filling tools. This proof-of-concept study shows that deep learning can be applied to the problem of gap-filling, and that at present, a complementary approach where GapPredict is used with other heuristic gap-filling approaches may be the best solution towards solving the de novo genome assembly gap-filling problem.
Acknowledgements:
This study was supported financially by the National Institutes of Health.
Image created with BioRender.com.
Learn more:
Learn more about the Birol Lab at the GSC and about their bioinformatics technology research
Learn about other bioinformatics tools and software for analyzing sequencing data
Citation:
Eric Chen, Justin Chu, Jessica Zhang, René L. Warren, Inanç Birol. GapPredict – A Language Model for Resolving Gaps in Draft Genome Assemblies. IEEE/ACM Transactions on Computational Biology and Bioinformatics.
*bold font indicates members of the GSC.