Using CRISPR-Cas9 technology, researchers from the Marra Lab generate CIC gene knockout immortalized human cell lines. Using their established cell lines, they assessed the transcriptomic and epigenomic changes following the loss of the CIC gene, which encodes Capicua, a transcriptional repressor protein implicated in neurodevelopment and oligodendroglioma.
Back to the basics: deleting a gene to determine its function
Gene deletion, also referred to as gene “knockout”, is a classical way to determine the function of the encoded protein. With the advent of CRISPR-Cas9 technology, knocking out genetic elements in cell lines has never been easier or cheaper to do in the laboratory.
Characterizing knockout cell lines to see how they differ compared to normal or “wild-type” cell lines can be done using various scientific techniques. For example, whole transcriptome sequencing (also known as RNA-Seq) can be used to see if the gene deletion results in the over- or under-expression of other genes (collectively, global gene expression, also referred to as RNA levels, is known as the “transcriptome”). Alternatively, you could check to see if epigenetic modifications (where all epigenetic modifications are referred to collectively as the “epigenome”) have been made following the deletion of your gene of interest.
Deleting a gene and noting the global impact of that deletion on gene expression (transcription) and epigenetic modifications provides clues on the function of the gene product (i.e., what does the protein encoded by that gene do within the cell?).
Capicua, a transcriptional repressor protein
Capicua (encoded by the CIC gene in humans) is a transcriptional repressor protein. By definition, a repressor protein is responsive to specific cues for the repression or silencing of target genes. The cues that guide Capicua function pertain to receptor tyrosine kinase (RTK) signalling: in the absence of RTK signals, Capicua is active and thus represses the transcription of its target genes; in the presence of RTK signalling, Capicua is inactive, thus target genes can be expressed (transcribed).
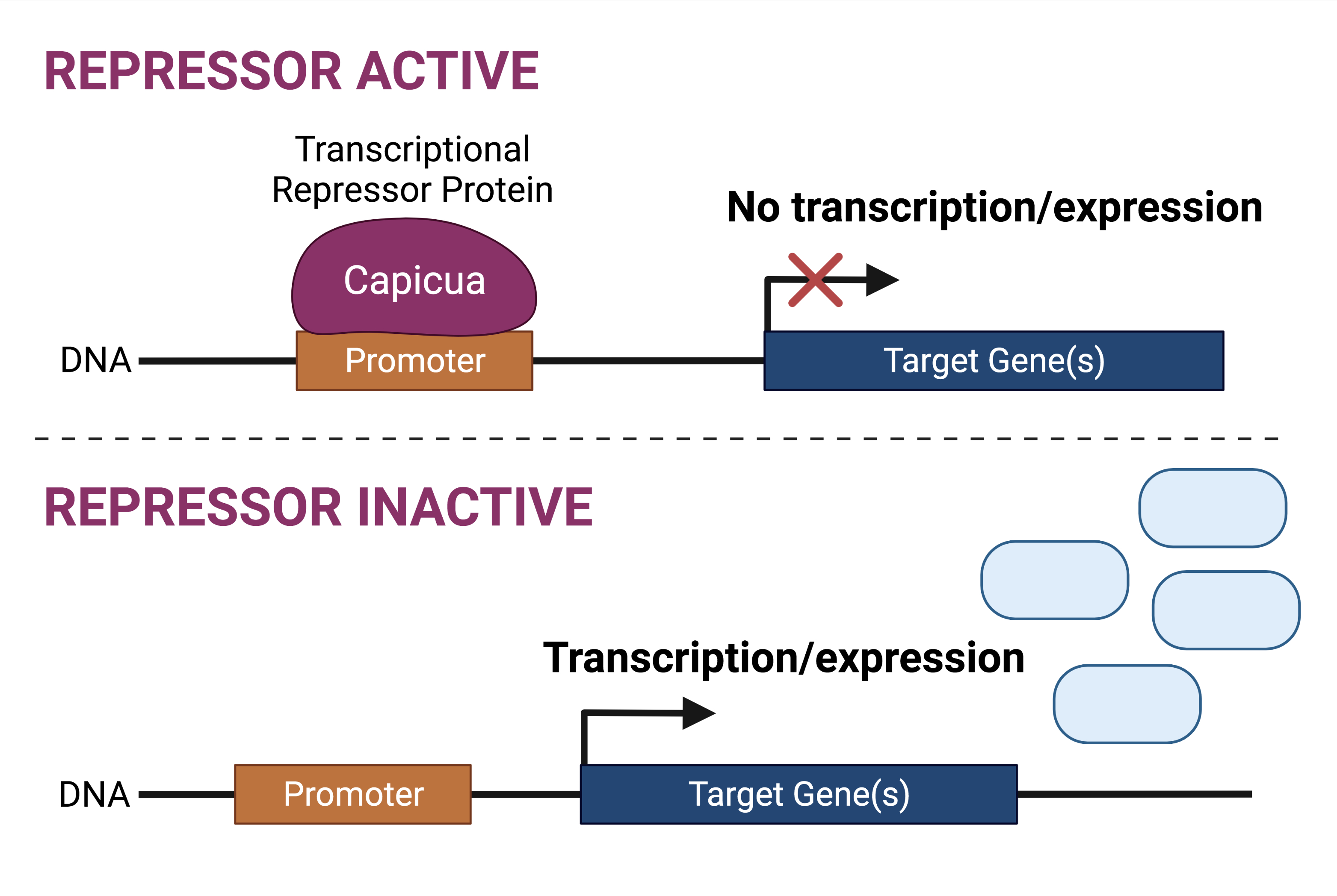
Previously, mutation of the CIC gene has been shown to impact neurodevelopment. In addition, mutations to the CIC gene have also been implicated in oligodendroglioma (tumour of the central nervous system) development, in conjunction with mutations to the IDH1/2 genes. There is some evidence that suggests the CIC gene may act in a tumour suppressive manner in oligodendroglioma.
Generating a knockout CIC cell line and characterizing its transcriptomic and epigenomic profiles
To learn more about the role the CIC gene plays in both processes, researchers from the Marra Lab used CRISPR-Cas9 technology to generate a CIC gene knockout in an immortalized human cell line and assessed genome-wide differences in the transcriptomic and epigenomic profiles between CIC wildtype and CIC knockout cell lines. Additionally, to better study CIC function in conjunction with the IDH1 gene, the researchers compared the transcriptome and epigenome of CIC knockouts with and without mutant IDH1 expression (the IDH1 gene is mutated to express R132H; where the original arginine amino acid at position 132 in the encoded protein is mutated to histidine instead). IDH1 mutations have previously been observed in oligodendroglioma tumours and may work in tandem with CIC mutations to dysregulate the normal cellular transcriptome or epigenome.
Their study, published in The Journal of Pathology, explores the functional implications of CIC loss, where the researchers were able to identify target genes involved in neurodevelopment and tumour development, including EPHA2 and ID1.
“This research highlights neurodevelopment and specific genes within this context as candidate targets through which CIC alterations may contribute to the progression of IDH-mutant gliomas,” said Stephen Lee, lead author on the study, and former GSC Master's student in the Marra Lab, now working as a Informatics Analyst at Zymeworks Incorporated. Lee committed to finishing this study even after leaving the GSC.
Acknowledgements:
This work was financially supported by the Canada Foundation for Innovation and the BC Knowledge Development Fund and the Canadian Institutes of Health Research Foundation Grants program.
Image created with BioRender.com.
Learn more:
Learn more about the Marra Lab at the GSC.
Learn more about CRISPR-Cas9.
Citation:
Lee SD, Song J, LeBlanc VG, Marra MA. Integrative multi-omic analysis reveals neurodevelopmental gene dysregulation in CIC-knockout and IDH1 mutant cells. J Pathol. 2021 Nov 12. doi: 10.1002/path.5835. Epub ahead of print. PMID: 34767259.
*bold font indicates members of the GSC.