In 2019, a novel, highly contagious coronavirus that caused respiratory illness began to spread globally, impacting millions of lives. Later, we would refer to this novel coronavirus as COVID-19 (COronaVIrus Disease 2019). Although the global pandemic is still currently ongoing, numerous groups have already conducted scientific research on predicting disease outcomes of COVID-19, including research efforts looking into the HLA alleles by the GSC’s Birol Lab.
The HLA gene family
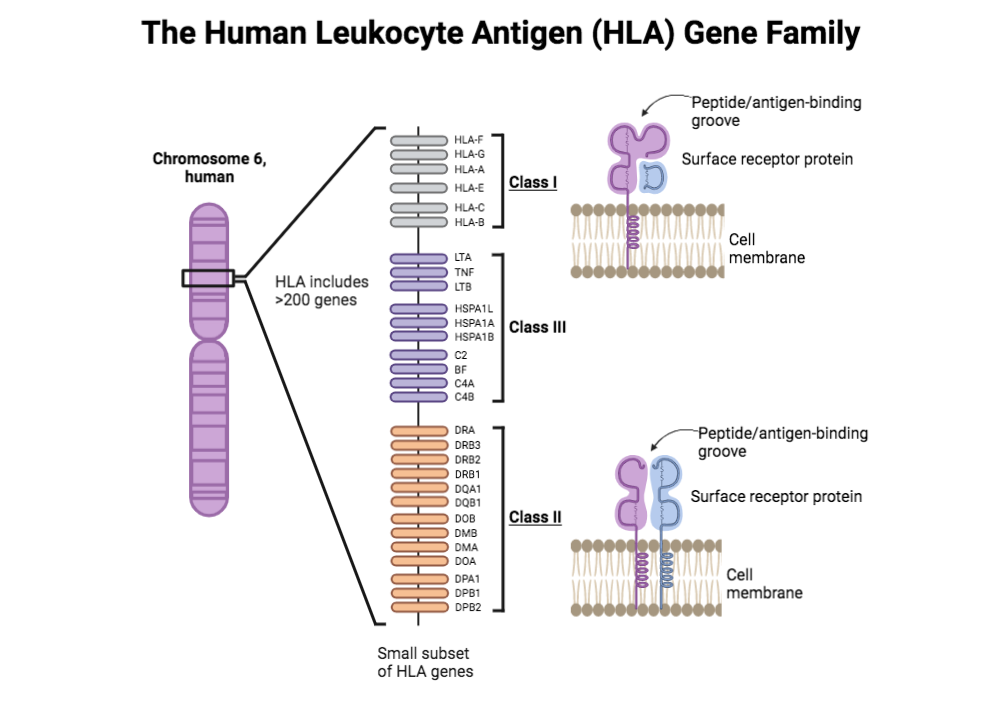
The human leukocyte antigen (HLA) gene family, also commonly referred to as the human version of the major histocompatibility complex (MHC), is a large group of genes located on chromosome 6. These genes, which can be divided into three classes (class I, II, and III), are essential for proper immune function and recognition of foreign pathogens by the body.
Many HLA genes code for cell-surface or receptor proteins that can be found on the surface of almost all human cells. These cell-surface proteins are capable of recognizing and binding specific, foreign peptides or “antigens”.
Between individuals, there is a high degree of disparity in terms of their immune response, and a major contributing factor to this diversity is the heterogeneity of the HLA system.
In the past, many scientists have conducted research looking at different HLA genes and their alleles to see if there is any association with disease. As published in the journal PeerJ, the Birol Lab has investigated possible associations between HLA and COVID-19 positive status, including disease severity.
COVID-19 outcomes
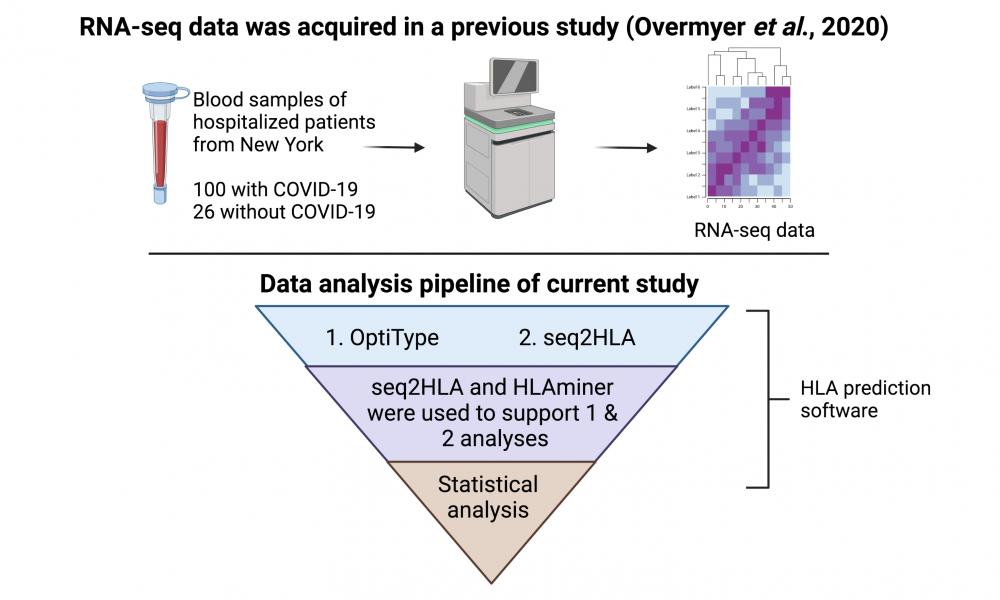
Previously, by analyzing publicly available metatranscriptomic sequencing data using HLA predictive software, the Birol Lab identified class I and II HLA genes and alleles, including DPA1*02:02 (a naming convention includes gene name*allele group/locus: specific HLA protein), by looking at two small samples of COVID-19 patients.
In the PeerJ study, using public RNA sequencing data with HLA predicative software, the researchers were able to analyze a larger cohort of COVID-19 patients with controls, and conducted further analysis to report on the association between HLA alleles and COVID-19 susceptibility and severity.
Similar to their previous findings, the researchers saw that DPA1*02:02 was observed at a higher frequency in the group of patients ill with COVID-19; however, having this allele alone did not correlate with increased risk of hospitalization.
Meanwhile, HLA-A*11:01 and, especially, HLA-C*04:01, were predominant in New York patients who needed intubation/mechanical ventilation, outcomes of a severe COVID-19 disease progression.
HLA alleles and COVID-19: a cautious association
The researchers caution that multiple considerations need to be considered when conducting genetic association studies. For example, the ethnicity and geographic location of the COVID-19 patients selected is important as populations can have varying allele frequencies (e.g., some populations may show a greater association than others).
Another important factor to consider is sample size, as smaller samples are more susceptible to variability. In the future, larger cohort sizes will be especially useful in assessing whether HLA alleles can be used as a prognostic tool for predicting COVID-19 outcomes.
Acknowledgements:
This study was supported financially by Genome Canada and Genome British Columba, and the National Institutes of Health.
Images created with BioRender.com.
Learn more:
Learn more about the Birol Lab at the GSC and about their bioinformatics technology research
Learn about other bioinformatics tools and software for analyzing sequencing data
Learn more about the Birol Lab's earlier HLA and COVID-19 study
Learn more about how the original RNA-seq data was acquired
Learn more about COVID-19
Citation:
René L. Warren, Inanç Birol. HLA Alleles Measured from COVID-19 Patient Transcriptomes Reveal Associations with Disease prognosis in a New York Cohort. PeerJ.
*bold font indicates members of the GSC.