Globally, COVID-19 has challenged healthcare systems. In addition to medical laboratory supply shortages, proprietary diagnostic reagents have also gone through periods of unavailability to the point where COVID-19 testing was impeded. To provide a sustainable supply of reagents, researchers from the GSC, BC Cancer Research Institute, University of British Columbia (UBC), UBC Michael Smith Labs, BC Centre for Disease Control (BCCDC) Public Health Laboratory, and PHSA Provincial Laboratory Medicine Services, collaboratively optimized a nucleic acid extraction protocol that used readily available reagents on an open-deck liquid handling robot, and was suitable for COVID-19 testing.
Isolating nucleic acids: columns vs. beads
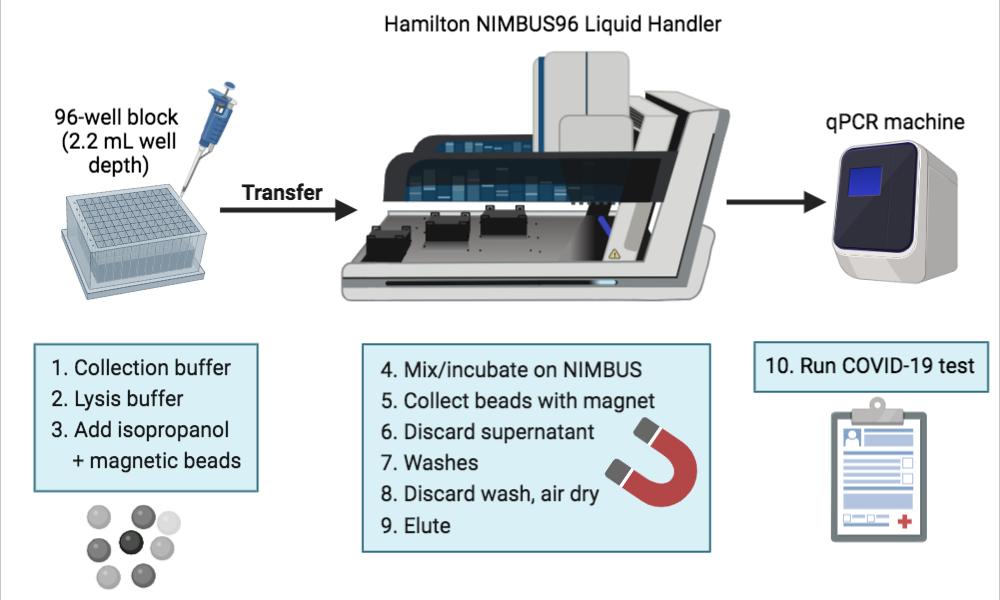
Extracting nucleic acids (e.g., DNA, RNA) involves two basic steps: first, breaking open or “lysing” cells to release their contents (e.g., proteins, DNA, RNA, etc.) and second, isolating (or purifying) the nucleic acids in the lysate using commercially manufactured purification columns, or magnetic beads. Both the columns and beads have been engineered such that they only bind and retain nucleic acids, while other lysate components can be washed away.
Column-based purification of nucleic acids relies on special filters or resins that bind the nucleic acids. Magnetic beads, which are coated by proprietary means to bind nucleic acids, are advantageous over columns as they can be more readily implemented in automated workflows, allowing for large-scale nucleic acid extraction and purification.
In addition to proprietary, commercially available kits, there are also protocols that use more economical and widely available magnetic beads and reagents to isolate nucleic acids from cells and tissue samples. Prior to the current study, these protocols had not been fully assessed for isolating viral nucleic acids suitable for clinical implementation.
Researchers thus sought to optimize and implement a high-throughput, bead-based nucleic acid isolation protocol, deployed on the readily available Hamilton Microlab NIMBUS robot, an open-deck 96-well liquid handling platform, for COVID-19 testing purposes.
Optimizing an automated open-deck liquid handling protocol

After multiple optimization and technical refinement trials, the new procedure was shown to exhibit specificity and sensitivity on par with a current, clinically-approved, proprietary COVID-19 testing approach. Final benchmarking tests were conducted with COVID-19 samples, and the researchers were able to achieve similar yield and comparable sensitivity and specificity using the newly developed protocol. Their research was published in the Journal of Virological Methods.
This work provides an alternative procedure for isolating nucleic acids from clinical samples using a generic protocol and reagents on an open-deck liquid handling platform. Proof-of-concept trials demonstrate that Standard Operating Procedure is as effective as the clinically-approved nucleic acids isolation protocol in terms of yield, sensitivity, and specificity. This new procedure serves as an excellent contingency plan to implement and alleviate the burden of potential diagnostic reagent shortages in the current pandemic and illustrates the innovation that is possible when a public health lab collaborates with a leading technology centre like Canada's Michael Smith Genome Sciences Centre.
Acknowledgements:
This work was supported by the Provincial Health Services Authority, Genome Canada and Genome British Columba, the Canada Foundation for Innovation and the BC Knowledge Development Fund.
Image created with BioRender.com.
Learn more:
Learn more about other technology development at the GSC, involving nucleic acids
Learn more about the GSC’s research into COVID-19, including its involvement with CGE to sequencing infected host genomes
Learn more about COVID-19 from the BC Centre for Disease Control
Learn more about the Hirst Lab’s research into COVID-19 testing
Learn more about the Hirst Lab, the Marra Lab, and the Jones Lab at the GSC
Citation:
Simon Haile, Aidan M. Nikiforuk, Pawan K. Pandoh, David D.W. Twa, Duane E. Smailus, Jason Nguyen, Stephen Pleasance, Angus Wong, Yongjun Zhao, Diane Eisler, Michelle Moksa, Qi Cao, Marcus Wong, Edmund Su, Martin Krzywinski, Jessica Nelson, Andrew J. Mungall, Frankie Tsang, Leah M. Prentice, Agatha Jassem, Amee R. Manges, Steven J.M. Jones, Robin J. Coope, Natalie Prystajecky, Marco A. Marra, Mel Krajden, Martin Hirst. Optimization of magnetic bead-based nucleic acid extraction for SARS-CoV- 2 testing using readily available reagents. Journal of Virological Methods.
*bold font indicates members of the GSC.