Hematopoietic differentiation—the creation of multiple blood cell types from one stem cell line—is complex and still poorly understood. Understanding how cells with different functions can arise from one “starter cell” is not only interesting, it is foundational to the development of disease treatment, including cancers such as leukemia. A new study from the lab of Dr. Martin Hirst sheds light on how a specific epigenetic modification signature contributes to the process.
Epigenetics is the study of how modifications to chromatin regulate gene activity to create different cell types from one genome. It is generally understood that epigenetics plays a role in hematopoietic differentiation, resulting in mature blood cells—including short-lived red blood cells—that are produced throughout a person’s lifetime. Research into how and when these epigenetics modifications occur is ongoing.
Reference Epigenomes from Cord Blood
A study from the Hirst Lab generated reference epigenomes for eight different cell types isolated from human cord blood. The resulting genome-wide datasets contain information on permissive (turning on gene expression) and repressive (turning off gene expression) histone modifications, sites of DNA methylation and transcriptomes.

“Our study presents the first reference epigenome profiles of primary CD34+ compartment of normal human cord blood, including the Human Stem Cell-containing CD38- subset and phenotypically distinguished subsets of the CD38+ cells,” says Dr. Alireza Lorzadeh, first author of the study and former PhD student in the Hirst lab, referring to progenitor or parent cells isolated from cord blood. “The low input assay I developed previously made it possible to generate these profiles without the intervention of in vitro expansion.”
H3K27me3 modification to Histone H3
Analysis of repressive chromatin states (areas of the genome where chromatin is condensed or tightly packed in a way that genes from that region cannot be expressed) revealed interesting results. Four of the progenitor cells shared a nearly identical epigenetic modification to histone H3 referred to as an H3K27me3 signature.
Past studies have associated repressive H3K27me3 chromatin modifications with regulation of hematopoiesis. In this study, the H3K27me3 signature seen in large areas of repressed chromatin in the progenitor populations was present in B and T cells but absent from monocytes and erythroblasts.
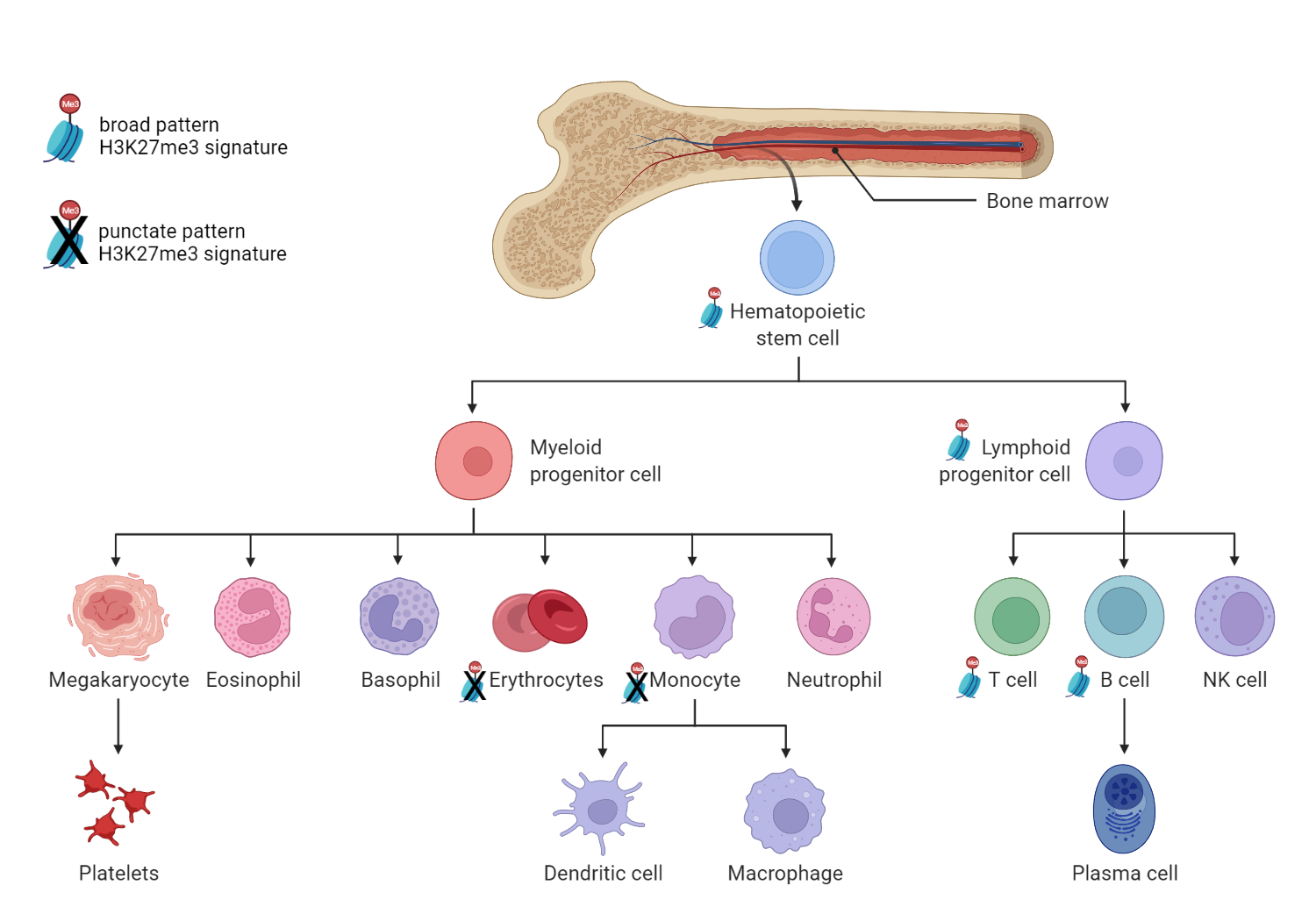
“We show that H3K27me3 reverts back to a 'pluripotent', punctate pattern in myeloid but not lymphoid lineages. This reinforces the concept proposed by many that H3K27me3 exists in two main states; broad and punctate,” says GSC’s Distinguished Scientist Dr. Hirst. “What is curious is that the H3K27me3 punctate pattern has been described as associated exclusively with pluripotency—but here we show that it is also found in terminally differentiating myeloid cells”
H3K27me3 Contraction
The team proposes a model based on these findings in which a genome-wide contraction of H3K27me3 signature is a critical step in the process by which human hematopoietic progenitor cells lose the ability to become lymphoid cells. Polycomb proteins—responsible for methylation of H3K27—are thought to play an important role in hematopoietic differentiation based on studies showing overexpression and inactivation of polycomb proteins in blood cancers.
“We are now testing the hypothesis that broad vs. punctate H3K27me3 occupancy is defined in part by higher order chromatin conformation changes,” says Dr. Hirst.
“This considerable resource of data and the findings derived from it has broad implications for the fields of hematological diseases as well as normal hematopoiesis,” says Dr. Lorzadeh, now a postdoctoral scholar in Dr. Jadhav’s Lab at the University of Southern California. “One of the main findings of this study is the striking contraction of H3K27me3 density, resembling that of embryonic stem cells, which has a predicted functional role in myelo-erythroid vs lymphoid lineage decision making in hematopoietic progenitor cells.”
Learn More:
Learn more about research in the Hirst lab and the Genome Science Centre.
Learn more about epigenetics and hematopoiesis.
Acknowledgements:
This work was supported by the Canadian Institutes of Health Research and Genome Canada, under the Canadian Epigenetics, Environment and Health Research Consortium, and by the Terry Fox Research Institute.
The authors thank the production and technical staff at Canada’s Michael Smith Genome Sciences Centre for generating human cord blood T cell reference epigenomes and their technical expertise in sequence generation and analysis. The authors also thank Darcy Wilkinson and Margaret Hale in the Eaves lab for collecting and processing CB cells, respectively.
Hematopoietic differentiation image created with biorender.com
Citation:
A. Lorzadeh, C. Hammond, F. Wang, D. J. H. F. Knapp, J. CH. Wong, J. Y. A. Zhu, Q. Cao, A. Heravi-Moussavi, A. Carles, M. Wong, Z. Sharafian, J. Steif, M. Moksa, M. Bilenky, P. M. Lavoie, C. Eaves, M. Hirst. Polycomb contraction differentially regulates terminal human hematopoietic differentiation programs. BMC Biology.
*bold font indicates members of the GSC.